The expression of epithelial mesenchymal transition related factors in human colorectal cancer cell lines
Article information
Abstract
Purpose:
Epithelial mesenchymal transition (EMT) is a well characterized embryological process thought to play a vital role in tumor progression. The purpose of this study was to evaluate the expression of EMT-related factors and then use EMT status to identify high metastatic potential in commonly used human colorectal cancer cell lines.
Methods:
In 11 commonly used cell lines, total mRNA expression levels were checked for the cell markers E- cadherin, vimentin, N-cadherin, and fibronectin, and the transcription factors snail, slug, twist, and SIP1 using real time PCR.
Results:
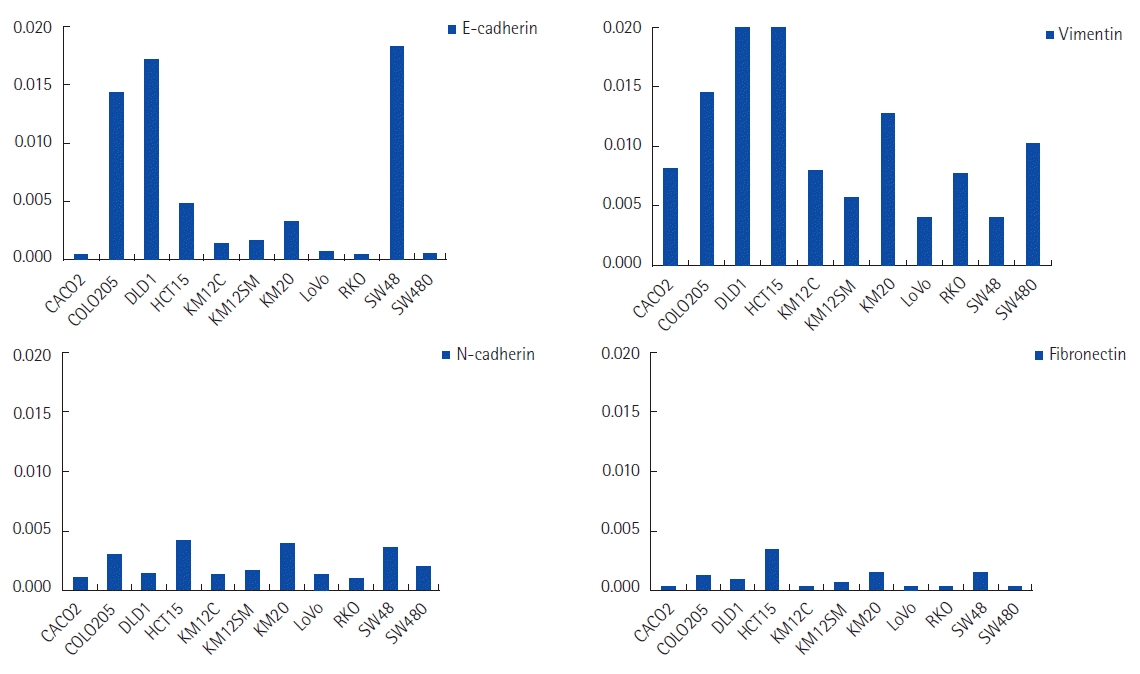
E-cadherin expression was positive in COLO205, DLD1, and SW48 cell lines. Vimentin expression was positive in COLO205, DLD1, HCT15, KM20, and SW480. Three different EMT status groups were defined according to mRNA expression of the cell markers. The epithelial phenotype expressed E-cadherin without vimentin (SW48), incomplete EMT phenotypes either concurrently expressed or did not express both E-cadherin and vimentin (CACO2, COLO205, DLD1, KM12C, KM12SM, LoVo, and RKO), and complete EMT phenotypes expressed vimentin without E-cadherin (HCT15, KM20, and SW480). The mRNA expression of transcription factors had no correlation with expression of vimentin, N-cadherin, and fibronectin, but there was an inverse directional correlation between mRNA expression levels of transcription factors and E-cadherin. There were no statistically significant correlations, however.
Conclusion:
Ten cell lines among the 11 were included in the incomplete and complete EMT groups. These findings could provide evidence of the high tumorigenesis and metastatic properties. The levels of mRNA expression and EMT status of human colorectal cancer cell lines can be applied to further investigations into cancer progression.
INTRODUCTION
Despite significant improvements in early diagnosis and treatment modalities, the mortality rates associated with invasive and metastatic cancer have not improved dramatically over time. Death from colorectal cancer results primarily from metastases [1], but the molecular regulation of cancer progression, including invasion and metastasis, is still unclear. A better understanding of the biological events that contribute to cancer progression is needed. It has been proposed that the same molecules that trigger the epithelial mesenchymal transition (EMT) are involved in tumor progression, including invasion and metastasis [2]. EMT consists of coordinated molecular and cellular changes defined as a loss of intercellular junctions, repression of the epithelial marker E-cadherin, and activation of the mesenchymal marker vimentin, resulting in a loss of epithelial polarity to disseminate and spread throughout the body [3]. These characteristics define the ‘complete’ EMT phenotype, which is correlated with cancer progression. However, ‘incomplete’ EMT subtypes that represent a mesenchymal phenotype also exist [4].
Recent research has documented the role of transcription factors in mediating the loss of epithelial adhesion molecules [3]. An inverse correlation between E-cadherin and snail expression has been noted in cultured cell lines established from various carcinomas [5,6]. The metastatic and tumorigenic potential of cell lines is usually higher than that of patient-derived tissues [7]. Moreover, cell lines offer advantages in preparation and handling that promote more accurate overall results. Therefore, for this preliminary work, we assessed the expression of EMT-related factors, including cell markers and transcription factors, in commonly used human colorectal cancer cell lines. The present study was attempted to classify the human colorectal cancer cell lines according to the EMT status. Furthermore, we tried to identify high metastatic potential in the cell lines through the analysis of EMT status.
METHODS
Cell lines and culture conditions
Eleven human colorectal cancer cell lines with well established characteristics were used [8-16] (Table 1). SW48 was obtained from the American Type Culture Collection (Rockville, MD, USA) and the rest were obtained from the Korean Cell Line Bank (Seoul, Korea). CACO2 and KM12C cells were grown in Minimum Essential Medium (MEM, Gibco-BRL, Paisley, UK) supplemented with 10% fetal bovine serum (FBS, Gibco-BRL), 3 g/L sodium bicarbonate (Sigma, St. Louis, MO, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco-BRL). COLO205, DLD1, HCT15, KM12SM, KM20, and LoVo cells were grown in RPMI 1,640 medium (Gibco-BRL) supplemented with FBS, penicillin, and streptomycin. RKO, SW48, and SW480 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco-BRL) supplemented with FBS, penicillin, and streptomycin. The growth patterns of the cell lines under conventional culture conditions were identified with an electron microscope without staining (phase optics, × 100).
RNA extraction
Total RNA from the cell lines was obtained using an RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. RNA concentrations were determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Montchanin, DE, USA), and 260/280 ratios ranged from 1.85 to 2.0.
Real time PCR
First-strand cDNA was generated by reverse transcription of 2 μg total RNA pre-sample with oligo dT primers using the Maxime RT PreMix Kit (iNtRON Biotechnology, Seoul, Korea) according to the manufacturer’s instructions in a final reaction volume of 20 μL. For real time PCR, 1 μL of cDNA was added to Power SYBR Green PCR master Mix (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions to achieve a final reaction volume of 10 μL (Table 2). Real time PCR was done using the 7900HT Fast Real-time PCR System (Applied Biosystems). The PCR protocol consisted of initiation at 50°C for 2 minutes and 95°C for 10 minutes for 1 cycle each, followed by amplification for 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The accuracy of the tests was checked by dissociation curve, and dissociation curves with two peaks were excluded from data collection. Ct data were collected via Sequence Detection Systems 2.3 software (Applied Biosystems). Real time PCR amplification of cell markers and transcription factors was done using the primer pairs listed in Table 2. E-cadherin which is the most representative cell marker of EMT, and three additional typically used mesenchymal markers were evaluated. Fold changes in target gene expression were normalized to GAPDH gene expression. We counted relative expression greater than 0.01 as positive.
Statistical analysis
Statistical correlations for identification of the relationship between E-cadherin and transcription factor expression were assessed by linear regression tests. Statistical significance was accepted for probability values less than 5% (P<0.05). All statistical analysis was performed using SPSS ver. 17.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
Growth patterns
Cell lines were classified into three groups based on their growth patterns (Fig. 1). All lines except COLO205 grew adherent to the base. RKO, SW48, and SW480 cells grew adherent to the base, but loosely attached. DLD1, HCT15, KM12C, KM12SM, KM20, and LoVo cells grew adherent to the base and strongly attached. COLO205 cells grew loosely attached in suspension. We found no differences in mRNA expression levels of cell markers based on growth pattern.
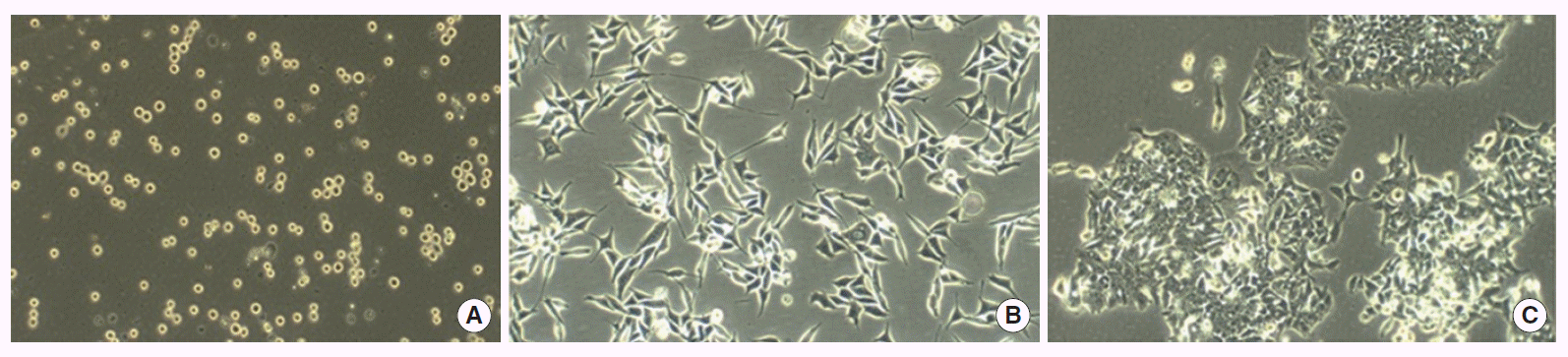
Growth patterns under conventional culture conditions. (A) Microscopic findings of in suspension loosely attached COLO205 cell line, (B) microscopic findings of adherent lossely attached RKO cell line, and (C) microscopic findings of adherent strongly attached LoVo cell line, all slides under phase optics (x100).
mRNA expression of cell markers
The relative mRNA expression of 4 different cell markers in 11 colorectal cancer cell lines was recorded (Fig. 2). COLO205, DLD1, and SW48 cell lines were positive for E-cadherin. Vimentin mRNA expression was positive in COLO205, DLD1, HCT15, KM20, and SW480 cells. All cell lines were negative for N-cadherin and fibronectin.
EMT status of colorectal cancer cell lines
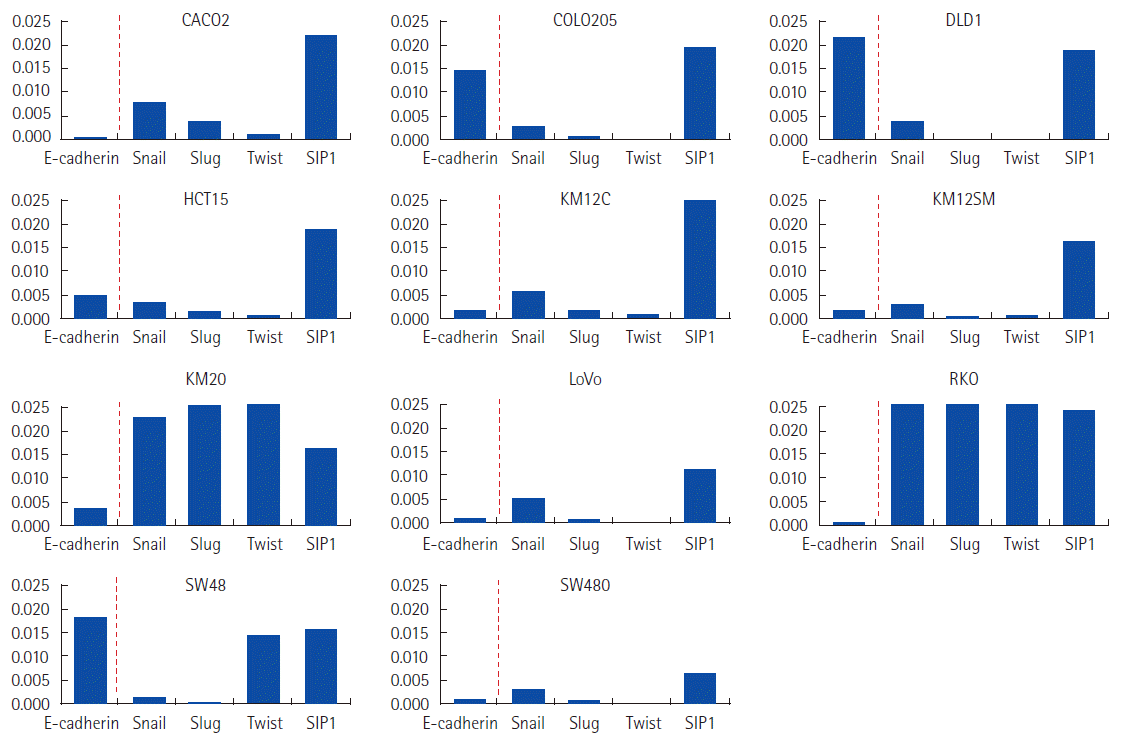
Three different groups of EMT status were defined according to mRNA expression of the four cell markers. The epithelial phenotype expressed E-cadherin without vimentin (SW48), incomplete EMT phenotypes either concurrently expressed or did not express both E-cadherin and vimentin (CACO2, COLO205, DLD1, KM12C, KM12SM, LoVo, and RKO), and complete EMT phenotypes expressed vimentin without E-cadherin (HCT15, KM20, and SW480) (Fig. 3). N-cadherin and fibronectin were excluded from the mesenchymal characterization because neither was detected in any cell line.
Relationship between E-cadherin and transcription factor expression
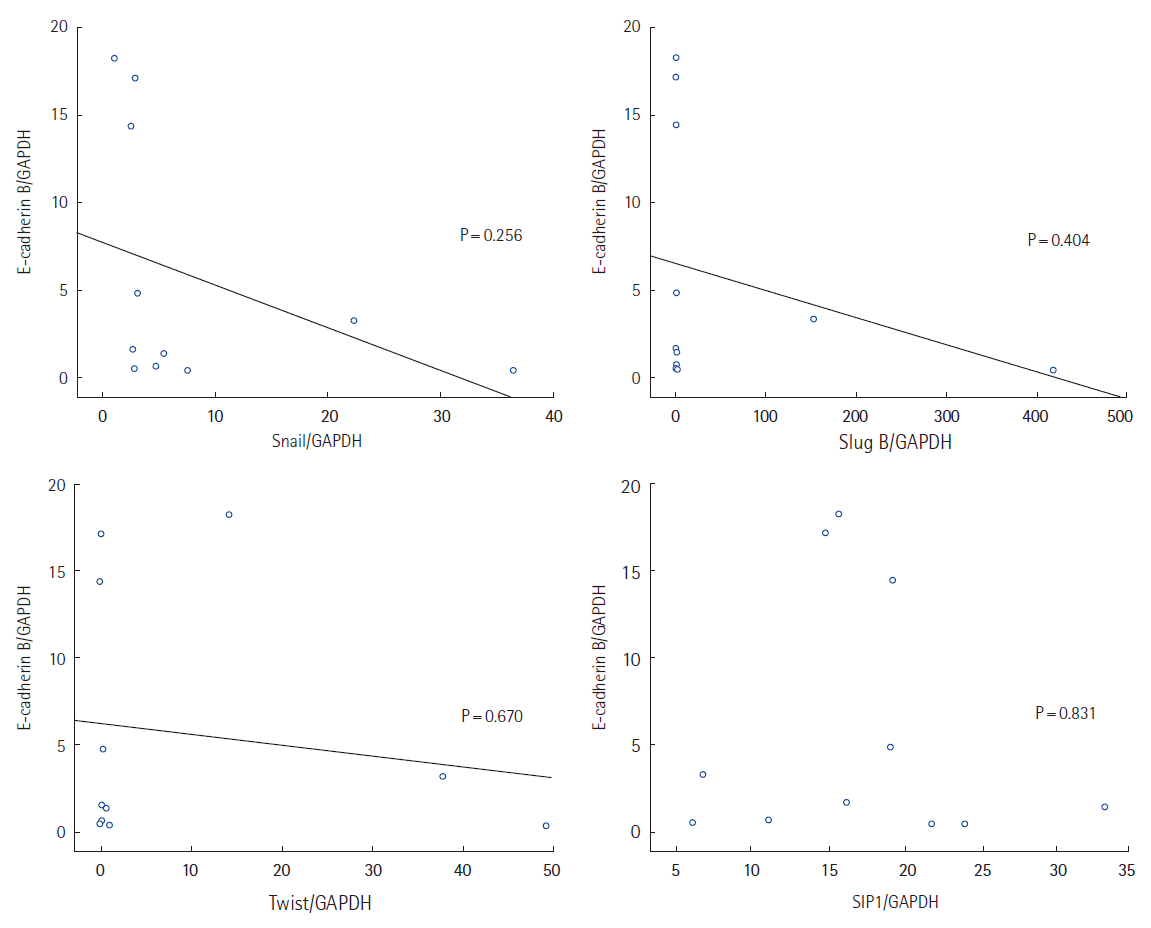
Possible correlations between mRNA expression levels of the four cell markers and four transcription factors were analyzed. There was no correlation between mRNA expression of transcription factors and vimentin, N-cadherin, or fibronectin. We found evidence of an inverse directional correlation between mRNA expression of the transcription factors snail, slug, and twist (but not SIP1) and the epithelial cell marker E-cadherin (Fig. 4), but none of the correlations were statistically significant (Fig. 5).
The correlation between the mRNA expression of E-cadherin and transcription factors. Y axis: mRNA expression level relative to the GAPDH primer.
DISCUSSION
The primary cause of death from colorectal cancer results from metastases that withstand conventional therapy . Carcinoma is the most frequent type of cancer in humans, and the occurrence of metastasis accounts for most cancer-related deaths. Invasion is the first of the cascade of events leading to development of metastasis, but it is at present the less understood. It occurs by transfer of malignant cells from the primitive neoplastic focus into surrounding host tissues and implicates acquisition of ability to migrate. The basic biological regulation of invasion and metastasis remains poorly understood [17], and cell lines offer advantages for research into cellular processes. Cell lines have higher tumorigenesis and metastatic properties than patient-derived tissues, and make it easier to accurately and reproducibly examine large mRNA pools. The relevant commonly used cell lines have been well characterized both in vitro and in vivo systems, including their invasive and metastatic properties [7]. In the present study, eleven human colorectal cancer cell lines with relatively well established characteristics were used. We observed typical growth patterns of each cell line, but did not identify any significant relationships between differences in growth patterns and the expression levels of cell markers.
EMT, first reported as a process of central differentiation in early embryogenic morphogenesis, is an essential step for numerous developmental processes, including mesoderm and neural tube formation [18]. EMT is a coordinated biological program characterized by a loss of cell adhesion, repression of E-cadherin expression, expression of N-cadherin or vimentin, and increased cell mobility. In the current study, we used the cell markers of EMT identified from several studies using other organ [19,20].
These characteristics define the ‘complete’ EMT phenotype. EMT plays a crucial role in the progression and aggressiveness of epithelial derived cancers [21]. Indeed several studies have confirmed the role of EMT in metastasis of colorectal carcinoma [22]. ‘Incomplete’ EMT subtypes that represent a mesenchymal phenotype also exist [5].
In this study, the mRNA expression levels of four cell markers and four transcription factors were recorded in human colorectal cancer cell lines. All tested cell lines had low mRNA expression of N-cadherin and fibronectin, making these markers inadequate for the characterization of colorectal cancer cell lines. We classified the cell lines into three groups through the analysis of mRNA expression levels of the cell markers (epithelial, incomplete EMT, and complete EMT phenotype), and found that EMT was relevant in 10 out of the 11 human colorectal cancer cell lines. These findings support the concept of high metastatic and tumorigenic potentials of these cancer cell lines.
An essential role for transcriptional factors in EMT has been demonstrated [23]. Transcription factors function as a direct repressor of E-cadherin, promoting EMT in several epithelial-derived cancers. An essential role for the transcriptional factors in EMT was demonstrated on the basis of its general induction in cellular conditions triggering this transition [24,25]. The transcription factors snail and slug may be related to colorectal cancer progression [26], and several recent reports have revealed connections between epithelial cancer progression and the transcription factors twist and SIP1 [27,28]. Likewise, the transcription factors snail, slug and twist regulate E-cadherin in breast cancer [29]. Slug also has been reported as a key gene promoting invasion in lung cancer [30]. In contrast with other recent studies, however, we did not find a significant correlation between E-cadherin expression and mRNA expression of transcription factors snail, slug, twist, and SIP1. We did observe an inverse directional correlation between the expression of transcription factors snail, slug, and twist and E-cadherin, but our numbers may have been too small to provide sufficient statistical power. According to our data, the transcription factors may play some role in EMT by down-regulating E-cadherin expression. No directional correlation was observed between expression of SIP1 and E-cadherin, indicating that SIP1 is inadequate for the study of EMT in colorectal cancer cell lines.
We identified mRNA expression of EMT-relatd cell markers and transcription factors in commonly used human colorectal cancer cell lines. Limitation of this study included that we did not find any differences in growth patterns according to the expression level of cell markers, and the expression of transcription factors was not significantly correlated with E-cadherin expression. However, the present study could provide basic information about the molecular regulation of colorectal cancer progression with EMT status, as a preliminary work. Loss of expression of epithelial markers and adhesion molecules including E-cadherin is important in cancer progression and development. Our findings do support the concept that cell lines have higher metastatic potential than patient-derived tissues, and suggest that the expression of EMT-related factors (cell markers and transcription factors) and the EMT status of human colorectal cancer cell lines can be applied to further investigations into cancer progression.
Notes
No potential conflict of interest relevant to this article was reported.
Acknowledgements
This study was supported by grants of the Korea Healthcare technology R&D project, Ministry for Health & Welfare Affairs, Republic of Korea (A092255).