See commentary "Integrating ctDNA and tumor tissue analysis in gastric cancer: a synergistic approach unveils prognostic insights" in Volume 19 on page 43. ABSTRACTPurposeCombined analysis of the variant composition of circulating tumor DNA (ctDNA) from cell-free plasma and DNA from tumor tissue could provide insight into the implications of the genetic alterations responsible for the intratumoral and intertumoral heterogeneity of gastric cancer. We aimed to evaluate the usefulness of this approach in these patients.
MethodsCell-free plasma and formalin-fixed paraffin-embedded tumor tissue samples from 46 patients with gastric cancer were examined. Targeted deep sequencing was performed using a commercially available kit.
ResultsThe cell-free DNA (cfDNA) concentration was higher in stage II–IV versus stage I patients and in larger versus smaller tumors. Only 12 of the 36 (33.3%) alterations in the tumor tissue samples were in concordance with those in the ctDNA samples. Two variants were in concordance in stage I samples and 10 in stage II–IV samples. Actionable variants that were detected in concordance were in the stage II–IV samples. Preoperative ctDNA positivity of actionable variants was significantly associated with cfDNA concentration, lymphatic invasion, N stage, and TNM stage. Cancer recurrence was significantly associated with tumor size, lymphatic/vascular invasion, TNM stage, and ctDNA-tumor tissue variant concordance.
ConclusionPreoperative ctDNA genetic analysis using a multigene panel offers substantial clinical benefits when performed in conjunction with targeted deep sequencing of tumor tissue. Concordance between preoperative ctDNA and tumor tissue mutations may serve as a prognostic indicator in patients with gastric cancer.
INTRODUCTIONGastric cancer (GC) is the fifth most common type of cancer globally, with 1.09 million new cases in 2020 and 1.77 million expected cases by 2040 [1]. Prognostic biomarkers for GC include microsatellite instability and human epidermal growth factor. Microsatellite instability correlates with a decreased risk of lymph node metastasis, tumor invasion, and mortality and a more durable response to immune checkpoint inhibitors. Human epidermal growth factor is associated with aggressive disease and reduced survival, and treatment of GC patients with trastuzumab significantly improves overall survival [2]. Therefore, these biomarkers are useful in both newly diagnosed and metastatic cases [3].
Recently adapted for clinical testing, tissue-based next-generation sequencing (NGS) detects several somatic genetic alterations targeted by chemotherapy and immunotherapy. However, owing to the high heterogeneity of advanced GC, it cannot account for spatial or temporal heterogeneity. To better understand the implications of genetic alterations for targeted therapy, analysis of plasma-derived circulating tumor DNA (ctDNA) has been suggested. In patients with advanced GC, high plasma concentrations of cell-free DNA (cfDNA) have been linked to peritoneal recurrence and poor prognosis [4–6] and trastuzumab resistance can be identified by monitoring of ctDNA clonal mutations [7]. However, the clinical benefits of preoperative ctDNA analysis have not yet been established for GC. In this study, we identified genetic alterations in ctDNA and matched tumor tissue samples and assessed the prognostic significance of ctDNA-tumor tissue concordance in patients with GC.
METHODSSample collectionA total of 46 patients with GC who had undergone gastrectomy at Seoul National University Bundang Hospital were enrolled in the study between September 2013 and April 2014. The participants provided written informed consents and the study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (B-1208-165-002). Blood samples were collected in ethylenediaminetetraacetic acid tubes at the operation room just before surgery and processed within 6 hours of collection. The samples were centrifuged for 10 minutes at 3,500 rpm, and the separated plasma was filtered using standard serum filters. The separated plasma and buffy coat were stored in a deep freezer in 1.5-mL microcentrifuge tubes. cfDNA was extracted from the separated plasma using a MagMaX Cell-Free DNA Isolation Kit (Thermo Fisher Scientific), and cfDNA levels were measured using a TapeStation High Sensitivity D1000 system (Agilent Genomics) in September 2017. DNA was extracted from the buffy coat using a QIAamp Blood Mini Kit (Qiagen). Surgically resected specimens were formalin-fixed and paraffin-embedded. Manual microdissection was performed to designate areas with a tumor cell percentage of approximately 60%. DNA was extracted from the designated areas using a QIAamp DNA FFPE Tissue Kit (Qiagen) and measured using a Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific). Clinical information were searched retrospectively in the patients’ electronic medical record.
Library preparation and NGSThe library was prepared using an Archer Reveal ctDNA 28 Concordance Kit (ArcherDX, Inc.), which combines the Archer VariantPlex (tumor tissue) and Reveal ctDNA 28 (ctDNA) kits. The assay uses an amplicon-based NGS approach to target hotspot regions in 28 genes commonly mutated in solid tumors (ALK, AKT1, AR, BRAF, CTNNB1, DDR2, EGFR, ERB2, ESR1, FGFR1, HRAS, IDH1, IDH2, KIT, KRAS, MAP2K1, MAP2K2, MET, NRAS, NTRK1, NTRK3, PIK3CA, PDGFRA, RET, ROS1, SMAD4, MTOR, and TP53). Archer assays use anchored multiplex polymerase chain reaction technology, enabling quantitation of the number of starting cfDNA molecules and error correction. Libraries were purified and sequenced using AMPure XP beads and a NextSeq 500 system (Illumina), respectively.
Data analysis and variant filteringData were analyzed using Archer Analysis Bioinformatics Platform 6.2 software. Variants with ≥5 alternate observations, ≥3 unique alternate observations, an allele frequencies outlier P-value ≤0.01, and a variant allele frequency (VAF) cutoff of 0.55% for ctDNA and 5% for tumor tissue were filtered in as true variants. Variants that were considered benign (those corresponding to synonymous, intronic, or non-coding regions or with a minor allele frequency of ≥0.01) or were also detected in the buffy coat samples were filtered out; these were considered as germline variants or noncancerous somatic variants such as clonal hematopoiesis variants. For the final variants considered as true-positive ctDNA and/or tumor tissue variants, the same variant detected in the counterpart sample indicated concordance at the read level, even when the variant filter-in criteria were not fully met.
Statistical analysisUnpaired t-test was used to compare the mean cfDNA concentration between stages. Univariate logistic regression was used to analyze the association of clinical characteristics with tier I/II variant detection and recurrence. Statistical analysis was performed with the IBM SPSS Statistics, version 29.0. Clinical characteristics significantly associated with tier I/II positivity or recurrence at the univariate level were included in a multivariate logistic regression model. Variants detected in both the tissue sample and matched ctDNA sample were considered concordant. Patients with no detectable variants in the tumor tissue were classified as non-concordant.
RESULTSThe median age of the 46 patients (30 men, 16 women) in this study was 64.5 years (range, 34–84 years) (Table 1). Twenty-two patients had stage I GC, and 24 had stage II–IV GC. Nine patients experienced recurrence within 2 years after diagnosis. The mean cfDNA concentration for all samples was 1.025 ng/μL (range 0.016–6.330), with significantly higher concentrations in stage II–IV samples (1.524 ng/μL) than in stage I samples (0.480 ng/μL, P=0.004) (Fig. 1). Tumor size ranged from 1.2 to 12.6 cm (median, 4.0 cm), and the cfDNA concentration was higher in cases with a tumor size higher than the median (1.413 ng/μL vs. 0.648 ng/μL, P=0.043).
The mean sequencing depths of the ctDNA and tumor tissue DNA were 899 and 484, respectively. Thirty-six genetic alterations in 13 cancer-related genes were detected in 22 plasma samples; none were detected in the remaining 24 samples. A median of one genomic alteration was identified per plasma sample (range, 1–8), and the most frequently mutated genes included TP53, KRAS, KIT, ERBB2, and IDH2 (Table 2). The VAF of these variants ranged from 0.13% to 4.22%. In the matching tumor tissue samples, 36 genetic alterations were detected in eight genes. Twenty-six tumor tissue samples had a median of one genomic alteration (range, 1–4), while the remaining 20 had none. The most frequently mutated genes in the tumor tissue samples were TP53, KRAS, and KIT. There were no significant differences in the mean number of variants in stage I versus stage II–IV patients in either the plasma or tumor tissue samples. In addition, 15 patients had no genomic alterations in either the ctDNA or tumor tissue samples.
Only 12 of the 36 alterations (33.3%) in the tumor tissue were in concordance with those in the ctDNA samples (Fig. 2). Moreover, only two variants in the stage I samples and 10 in the stage II–IV samples were in concordance.
All variants were classified into four tiers based on their level of clinical significance in terms of cancer diagnosis, prognosis, and/or therapeutics, according to the recommendations of the proposed guideline; tier I, variants with strong clinical significance; tier II, variants with potential clinical significance; tier III, variants of unknown clinical significance; and tier IV, variants deemed benign or likely benign [8]. Variants that were higher than tier II were filtered. There were 12 tier II ctDNA variants and 27 tier II tumor tissue variants, with seven showing concordance (Fig. 3). There were no concordant variants among the stage I samples. In contrast, seven of the 17 tier II variants (41.2%) in the stage II–IV tumor tissue samples were concordant with those in the ctDNA samples. Interestingly, five tier II variants were only detected in the ctDNA samples of four patients (two, stage I; two, stage III; and one, stage IV) (Table 3).
To assess the association of the preoperative positivity of tier II variants with additional clinical factors, binary logistic regression was performed (Table 4). ctDNA positivity of tier II variants significantly correlated with higher cfDNA concentration, lymphatic invasion, N stage, and TNM stage II–IV in the univariate analysis; however, none of these associations remained significant in the multivariate analysis. Tier II positivity of tumor tissue samples significantly correlated only with sex, with an odds ratio for women of 5.18 (95% confidence interval, 1.34–20.06; P=0.017).
Because cancer recurrence was unrelated to tier II positivity in ctDNA and tissue samples, we sought to identify factors that were related. Univariate logistic regression showed that tumor size, lymphatic invasion, vascular invasion, T-stage, N-stage, M-stage, and tumor tissue-plasma variant concordance were significantly associated with cancer recurrence, whereas age, sex, and cfDNA concentration were not associated (P>0.05) (Table 5). However, in the multivariate model, none of the associations were significant.
DISCUSSIONIn this study, we analyzed the plasma ctDNA and tumor tissue DNA of 46 GC patients using a sequencing panel with the same target regions for each. We also extracted DNA from the buffy coat to filter out variants that did not originate in the tumor. Higher cfDNA concentration in the plasma samples correlated with higher stage and larger tumor size. However, plasma cfDNA alone remains insufficient as a clinical biomarker because of its lack of specificity [9]. The number of ctDNA variants in the plasma did not significantly differ between tumor stages or tumor sizes.
At least one genetic variant was identified in each of the ctDNA samples of 22 patients (22/46, 47.8%), with a VAF ranging from 0.13% to 4.22%. Only one-third of these variants were also detected in the tumor tissue samples. Our detection and concordance rates appear to be lower than those reported in previous studies on ctDNA analysis in GC patients [10,11]; this may be due to the exclusion of possible clonal hematopoiesis variants in our study. Concordant variants were more frequently detected in stage II–IV patients than in stage I patients. Among the potentially actionable tier II variants, seven (all from stage II–IV samples) showed ctDNA-tumor tissue concordance. In non-small cell lung cancer, higher input concentrations of cfDNA and later stage were associated with higher concordance [12]. Our data show that stage II–IV GC patients have higher cfDNA concentrations, as well as higher concordance, than do stage I GC patients, even though the number of tumor tissue variants did not differ significantly.
Although two-thirds of the total variants were not concordant, the top three most frequently mutated genes were present in both the ctDNA and tumor tissue samples (TP53, KRAS, and KIT). KRAS and KIT variants were detected more frequently in the present study than in previous studies [13,14]. Five tier II variants were identified only in the ctDNA samples (three KRAS variants in two stage III–IV patients and two TP53 variants in two stage I patients); the VAF of these variants ranged from 0.65% to 1.29%. These findings suggest that the number of total variants identified in conventional tissue biopsies may be underestimated owing to tumor heterogeneity. The ability to identify multiple concurrent heterogeneous resistance mechanisms is an advantage of ctDNA analysis. In a previous study in which 34 patients with gastroesophageal adenocarcinoma underwent baseline triplet-paired sequencing of primary tumors, metastatic tumors, and plasma ctDNA, 32% of the genetic variants in were concordance [13]; ctDNA-primary tumor concordance was 37%, while ctDNA-metastatic tumor concordance was 43%, suggesting that ctDNA analysis may more effectively guide targeted therapy selection for metastatic tumors than for primary tumors. In the present study, false-positive results were possible because the mean depth may have been insufficient for achievement of a low VAF, and tissue variants may have been missed in minor fractions with a VAF of <5%.
In our study, preoperative positivity of tier II ctDNA variants was associated with a high cfDNA concentration, lymphatic invasion, and tumor N stage. These observations are in line with a previous report showing that advanced disease and lymph node involvement increase the likelihood of detectable preoperative ctDNA[15]; however, lymphatic invasion was unrelated to preoperative ctDNA positivity in other studies [16,17]. In our analysis of GC patients, preoperative ctDNA positivity was not associated with recurrence, as it was in patients with lung cancer [18], with the caveat that it likely correlated with advanced disease rather than simply predicted recurrence in the lung cancer patients [19]. When we examined potential predictors of recurrence using preoperative data, variant concordance between ctDNA and tumor tissue significantly correlated with recurrence, along with tumor size, lymphatic invasion, vascular invasion, and stage. This finding suggests that combined preoperative ctDNA and tissue genetic profiling have complementary benefits in terms of predicting recurrence. This premise should be further evaluated in a larger cohort, since only nine patients with recurrent cancer were included in the present study. Large prospective studies are required to validate the clinical utility of preoperative ctDNA testing for GC [20]. Unfortunately, not being able to analyze the ctDNA after the recurrence would be a limitation of this study.
In conclusion, preoperative ctDNA genetic analysis using a multigene panel offers substantial clinical benefits when performed in conjunction with tumor tissue NGS. Concordance between preoperative ctDNA and tumor tissue mutations may serve as a prognosis indicator in patients with GC.
NotesFUNDING
This study was supported by the Seoul National University Bundang Hospital Research Fund (grant number 14-2014-0014).
REFERENCES1. Global Cancer Observatory. Estimated number of new cases from 2020 to 2040, both sexes, age [0-85+] [Internet]. International Agency for Research on Cancer;c2020. [cited 2023 Nov 28]. Available from: https://gco.iarc.fr/tomorrow/en/dataviz/isotype?cancers=7&single_unit=50000.
2. Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med 2019;25:1415-21.
3. National Comprehensive Cancer Network (NCCN). Gastric Cancer (Version 2, 2022) [Internet]. NCCN;c2022. [cited 2023 Nov 28]. Available from: https://www.nccn.org/professionals/physician_gls/pdf/gastric.pdf.
4. Fang WL, Lan YT, Huang KH, Liu CA, Hung YP, Lin CH, et al. Clinical significance of circulating plasma DNA in gastric cancer. Int J Cancer 2016;138:2974-83.
5. Nakamura Y, Kawazoe A, Lordick F, Janjigian YY, Shitara K. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: an emerging paradigm. Nat Rev Clin Oncol 2021;18:473-87.
6. Lengyel CG, Hussain S, Trapani D, El Bairi K, Altuna SC, Seeber A, et al. The emerging role of liquid biopsy in gastric cancer. J Clin Med 2021;10:2108.
7. Wang Y, Zhao C, Chang L, Jia R, Liu R, Zhang Y, et al. Circulating tumor DNA analyses predict progressive disease and indicate trastuzumab-resistant mechanism in advanced gastric cancer. EBioMedicine 2019;43:261-9.
8. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017;19:4-23.
9. Huang ZB, Zhang HT, Yu B, Yu DH. Cell-free DNA as a liquid biopsy for early detection of gastric cancer. Oncol Lett 2021;21:3.
10. Varkalaite G, Forster M, Franke A, Kupcinskas J, Skieceviciene J. Liquid biopsy in gastric cancer: analysis of somatic cancer tissue mutations in plasma cell-free DNA for predicting disease state and patient survival. Clin Transl Gastroenterol 2021;12:e00403.
11. Wallander K, Eisfeldt J, Lindblad M, Nilsson D, Billiau K, Foroughi H, et al. Cell-free tumour DNA analysis detects copy number alterations in gastro-oesophageal cancer patients. PLoS One 2021;16:e0245488.
12. Jiang J, Adams HP, Yao L, Yaung S, Lal P, Balasubramanyam A, et al. Concordance of genomic alterations by next-generation sequencing in tumor tissue versus cell-free DNA in stage I–IV non-small cell lung cancer. J Mol Diagn 2020;22:228-35.
13. Maron SB, Chase LM, Lomnicki S, Kochanny S, Moore KL, Joshi SS, et al. Circulating tumor DNA sequencing analysis of gastroesophageal adenocarcinoma. Clin Cancer Res 2019;25:7098-112.
14. Nakamura Y, Taniguchi H, Ikeda M, Bando H, Kato K, Morizane C, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med 2020;26:1859-64.
15. Yang J, Gong Y, Lam VK, Shi Y, Guan Y, Zhang Y, et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis 2020;11:346.
16. Kim YW, Kim YH, Song Y, Kim HS, Sim HW, Poojan S, et al. Monitoring circulating tumor DNA by analyzing personalized cancer-specific rearrangements to detect recurrence in gastric cancer. Exp Mol Med 2019;51:1-10.
17. Lee CS, Kim HS, Schageman J, Lee IK, Kim M, Kim Y. Postoperative circulating tumor DNA can predict high risk patients with colorectal cancer based on next-generation sequencing. Cancers (Basel) 2021;13:4190.
18. Isaksson S, George AM, Jonsson M, Cirenajwis H, Jonsson P, Bendahl PO, et al. Pre-operative plasma cell-free circulating tumor DNA and serum protein tumor markers as predictors of lung adenocarcinoma recurrence. Acta Oncol 2019;58:1079-86.
Fig. 2Number of variants detected in plasma circulating tumor DNA (ctDNA) and tumor tissue samples. (A) Variants detected at all stages. (B) Variants detected in stage I patients. (C) Variants detected in stage II–IV patients. 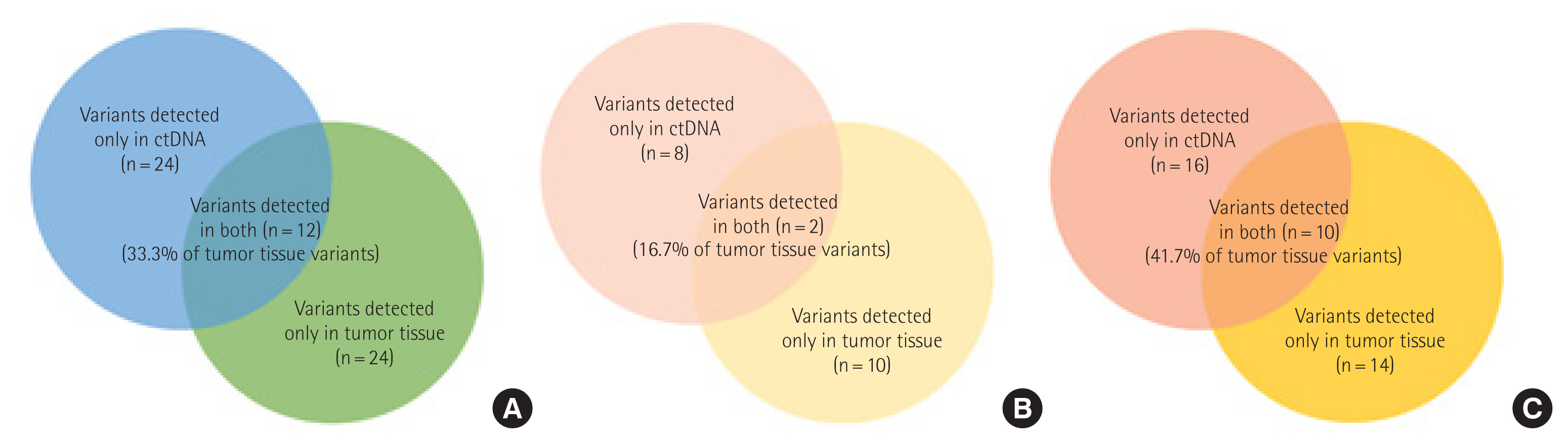
Fig. 3Number of tier II variants detected in plasma circulating tumor DNA (ctDNA) and tissue samples. (A) Tier II variants detected at all stages. (B) Tier II variants detected in stage I patients. (C) Tier II variants detected in stage II–IV patients. 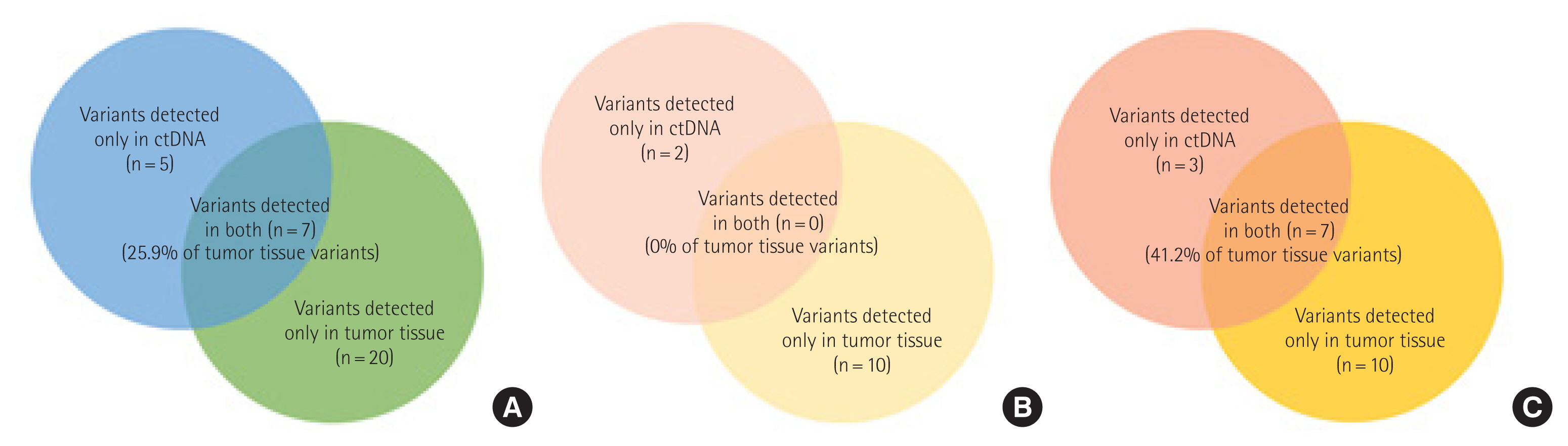
Table 1Patient characteristics
Table 2Frequently mutated genes in tumor tissue and ctDNA Table 3Tier II variants detected only in ctDNA Table 4Predictors of ctDNA positivity of tier II variants
Table 5Predictors of recurrence in the univariate analysis
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||